Contributing an Entry on SKY: A Tutorial
This article describes how to contribute an entry on SKY (consensus article, individual research report, methodological guideline). For contributing an entry to the open source book “Statistical Tools for Causal Inference”, see here.
SKY uses RMarkdown and Git. Awesome broad tutorials for these two softwares are:
You do not need to read the two books to use SKY. But as you become more proficient, most of your questions will be answered in these books.
Installation
Overall software
In order to get SKY up and running on your computer, you have to:
- Install R.
- Install Rstudio.
- Install Git.
- Create an account on GitHub. The entire procedure is described here.
Rstudio is a GUI, so you could manage without it, but at a tremendous cost. Rstudio features amazing tools to help process RMarkdown code. It is also integrated with Git, which makes the SKY experience almost seamless. You won’t have any command to drop in the shell :( Don’t worry, if you really want to feel like a true hacker, you can use the Git command line ;)
You could also manage without a GitHub account, but that would be dreary.
Packages
In R, you want to install the following packages:
- metafor: for running meta-analysis
- RMySQL: for dialoguing with the MySQL database.
- gsheet: for dialoguing with Google Docs.
- ggplot2: for beautiful graphs.
- dplyr: for easy data manipulation.
- knitr: for running the RMarkdown code.
In order to install these packages, do as follows:
- Open Rstudio
- Click on
Tools,Install Packages - Enter the names of the packages in the dialog box.
Or go to the Packages panel on the bottom right corner of the Rstudio window and click on Install.
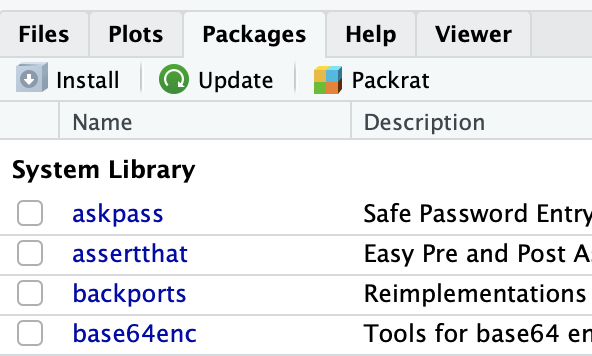
In the window that has just opened, enter the name of the packages separated by either a space or a comma.

Done!
Getting started
You’re almost almost there.
Forking SKY on GitHub
Yep, that does not sound super cool, but it’s not as bad as it sounds. It just means that you are going to create your own version of SKY on GitHub. It is not necessary (you could dowload SKY directly from my repository), but it makes interactions much smoother because we can use all the GitHub infrastructure to communicate and merge your changes to SKY, instead of resorting to lines of code, and everyone hates lines of code, right? Convinced?
OK, so you just have to log in to GitHub and to go to the original GitHub repository. Now, just click on the Fork button that you can see on the top right corner.

After a few seconds, you should be directed to your own repo, with your own copy of SKY.
Downloading SKY
In order to download SKY on your computer, follow the following steps in Rstudio:
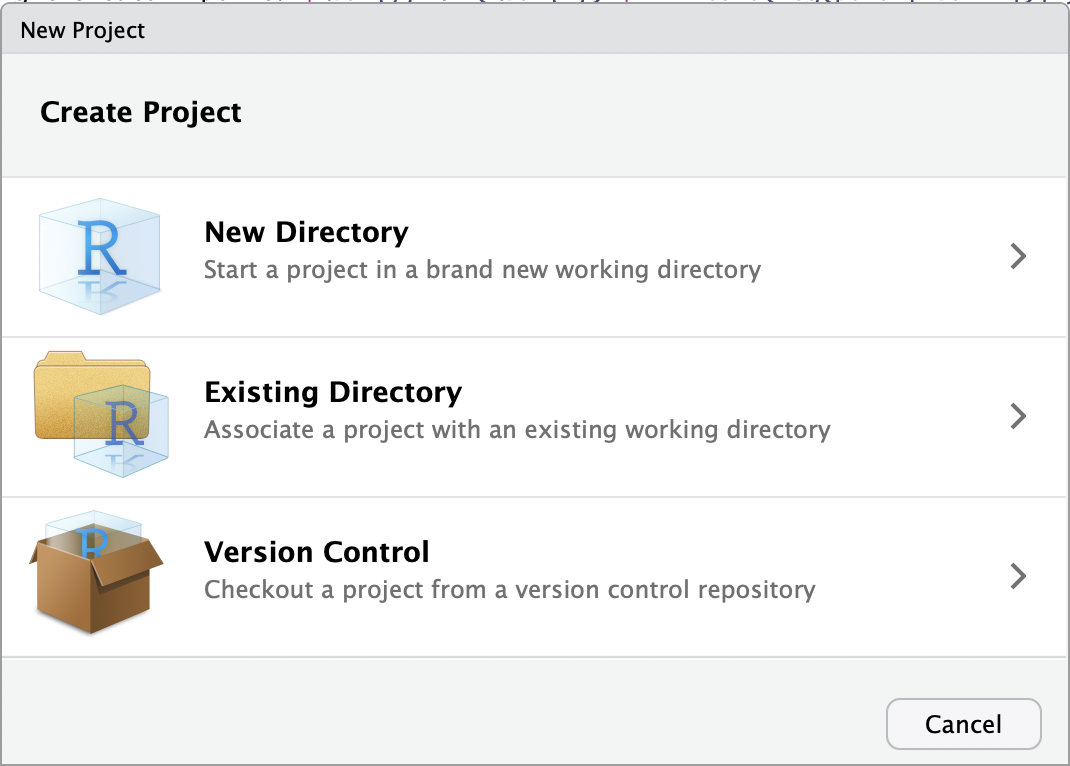
- Click on
File,New Project... - In the dialog box, click on
Version Control
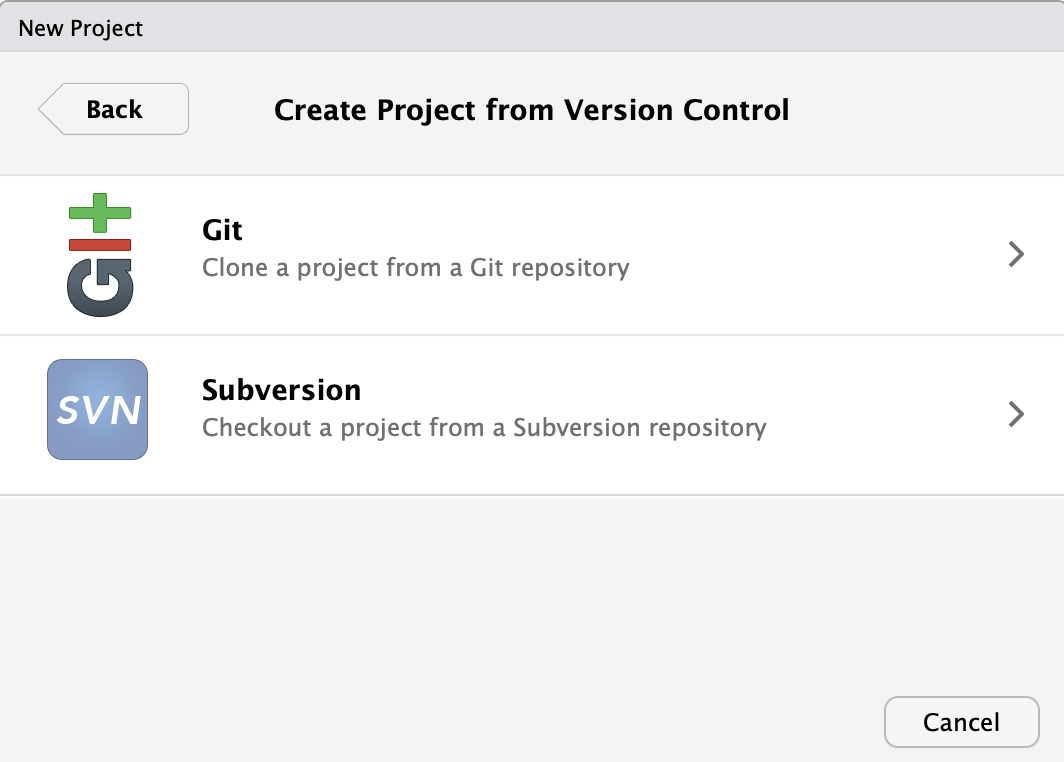
- In the following dialog box, click on
Git
- In the following dialog box, enter
https://github.com/YourGitHubUserName/SKYin theRepository URLfield, whereYourGitHubUserNameis your GitHub username.
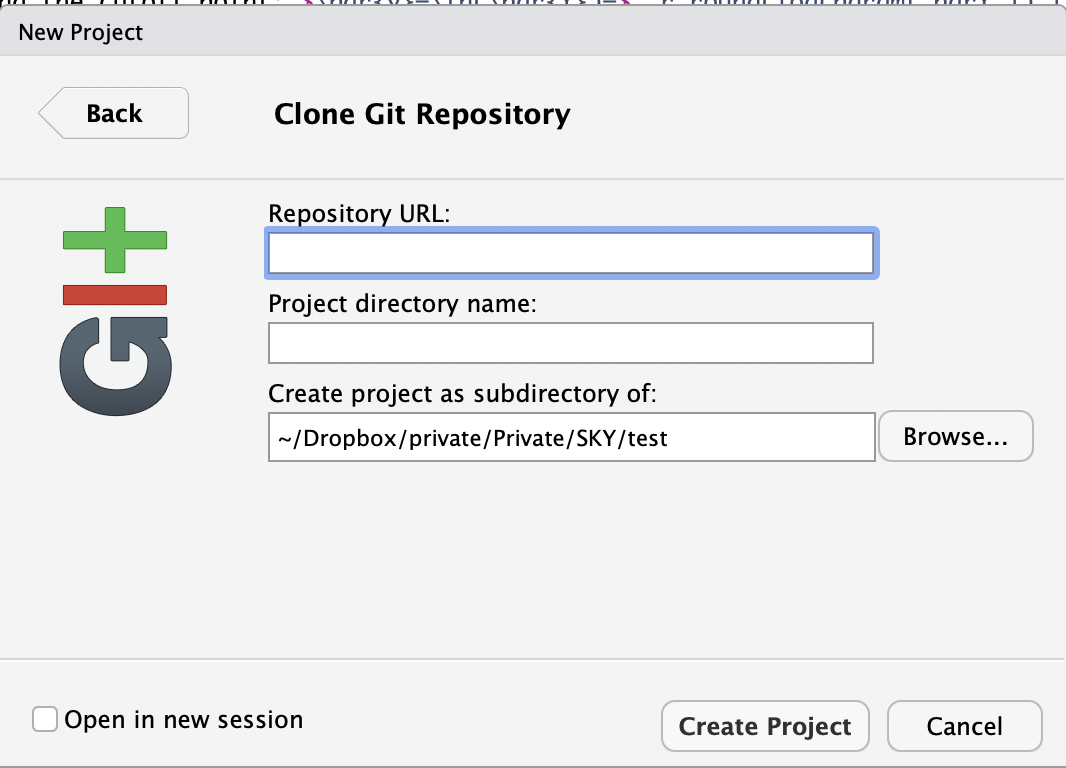
If you’re not sure what your username is, just copy/paste the whole address of the repo from the address bar of your web browser.
- Choose any name you want for the directory, although SKY seems appropriate.
- Click on
Browseto select where you want the project to be on your computer. - Don’t check the
Open in new sessionbox unless you wish to keep the current session open while a new Rstudio session is created with SKY on it. - Click on
Create Project
Rstudio clones all the files from the SKY repository and opens a session with SKY as main project. You’re almost there.
Id and password
Before hitting the Build button (I know you want to :), there is one extra step (otherwise, you’re gonna have an error message and that’s really depressing). You need to receive an Id and a password for accessing the MySQL database (I cannot grant access to everyone for now). In order to do so:
- Drop an email requesting id and password for the SKY SQL database at SKY.initiative.SCF[at]gmail.com
- Save the
source_blank.Rfile assource.R. Do not forget this last step, otherwise everyone in the world will have access to your id and password for the SKY MySQL database). - Update the username and password you’ve just received into the
source.Rfile, so that it reads:
username <- 'yourusername'
password <- 'yourpassword'with yourusername and yourpassword the username and password that I’ve just sent you.
Build
OK, now it’s time to hit the Build button and to get SKY to run on your computer. In order to do so, let’s click on the Build panel that is normally on the top right corner of the Rstudio window.
Go ahead, and hit the Build Website button, yes, the one with a nice hammer on it.
Shazam, you should have all the html files generated as well as a viewer opening with the SKY website there for your eyes to see.
Making changes to SKY
OK, so now, you’re in the big leagues, you know how to do collaborative open science. How to do it in practice? Here goes.
Now you can make all the changes you want to SKY. You can update the text. You can tinker with the code. You can add a new webpage. You can add new data. Here is how you would do that.
Updating the text
Updating the text is the easiest thing to do on SKY. Here is how to do it:
- Locate the name of the page that you want to update. For example, go on SKY’s webpage and locate a page. Or, click on the
Build Websitebutton, go to the Rstudio Viewer (it should open automatically), click on theOpen in Browseron the top left corner of the Viewer. Navigate the website in the browser (you’re actually navigating your own version of the website). Look at the url of the page in the address bar of your browser. What you’re looking for is the last part of the url: the one after the last/and before the.html. For example, for the urlhttps://chabefer.github.io/SKY/GrasslandPES.html,GrasslandPESis the name of the page. - Open the RMarkdown file that has generated this page. To do so, go to the
Filespanel in the bottom right corner of the Rstudio window.
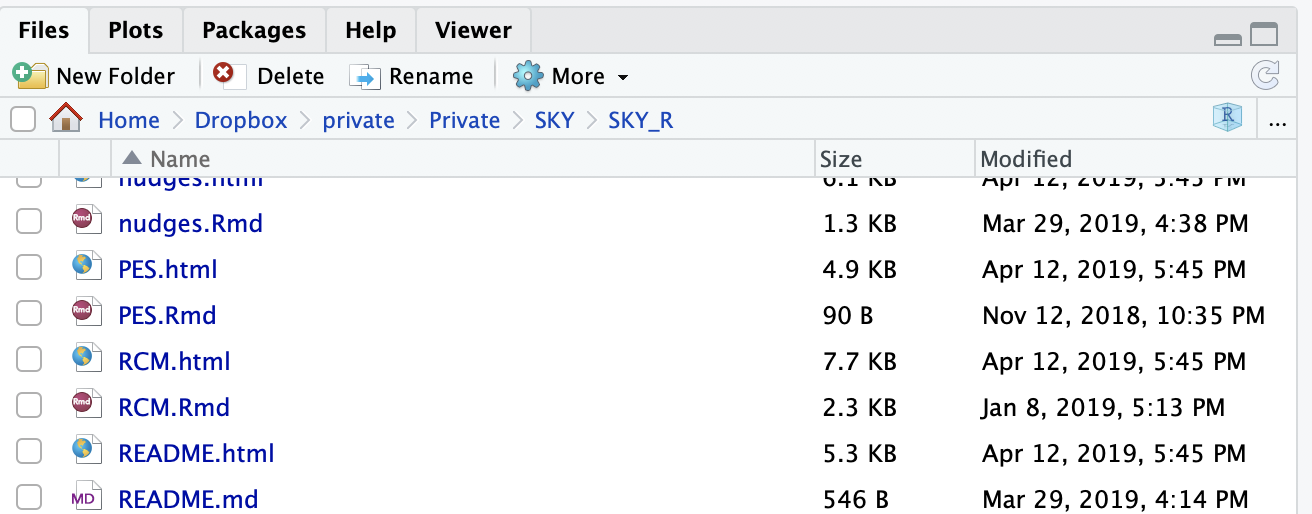
Locate the file that has the same name as the page you want to update but ends with .Rmd. It should be easy to do, since the files should be organized alphabetically. If they are not, you can make them by clicking on Name. For example, you can locate the file GrasslandPES.Rmd, or the .Rmd file behing the page that you’re reading right now, tutoSKY.Rmd. 3. Make changes to the text in the Rmarkdown file. In order to do make changes, you need to understand a little about the Markdown syntax. All you need to know to get started is here.
Tinkering with the R code
What is awesome with an Rmarkdown document is that the code analyzing the data is seamlessly integrated in the same .Rmd file as the text commenting it, so that each time you build the website, or Knit the file, the R code is run and tables, figures, etc, are automatically generated. That’s what makes the constantly updated meta-analysis possible. Each change to the code and data is seamlessly translated in the web page or the pdf document. Also, you never lose track of how a results has been generated, so that it is costless to check a results by yourself. And if you do not want the code to run each time, the amazing cache option enables you to store the results from the chunk and neveer compute them as long as the code chunk has not changed. Pretty cool, huh?
R code chunks appear in grey in between ticks in the .Rmd files. For example, the fourth code chunk in the GrasslandPES.Rmd file reads:
```{r constants,echo=FALSE,results=FALSE,warning=FALSE,error=FALSE,message=FALSE}
SCC <- 24
``````, three ticks, marks the beginning and end of a code chunk. I’ll come back to code chunks later. But for now, you can locate each part of the R code and make changes to the statistical analysis.
Adding a new webpage
Now, you want to create a webpage for your own project. How would you do that? Well, read on.
Click on the + button on the top left corner of the Rstudio window.

Click on R Markdown and follow the steps (nothing irreparable is done here, so you can just keep the defaults settings). Save the new page under the name you want to give it. Remember, this name will be the url address of your page, so choose wisely. I suggest a short name, with some initials and a clear link to the goal of the page. Also, the page name has to be new, please check in the list of files that the name you have in mind has not already been attributed.
Now, you’re done. You can start playing around with the page, changing the title, writing text, adding code.
Do not forget to link your new page to the rest of SKY, either by linking it directly on the front pages Interventions and Theories, or by linking to another article in SKY. The RMarkdown command to generate a link is simply [text](NameOfYourPage.html).
Adding new data
There are several ways to bring data to SKY. All of them are illustrated in the GrasslandPES.Rmd file.
The first way is simply to link to a GoogleSheet where estimates are stored or papers are listed. In order to do so, hit the Share button on the GoogleSheet page, choose Anyone with the link can edit and copy/paste the link to SKY using the Rmarkdown command [text](link). See the example just above the code chunk sendingGCP in the GrasslandPES.Rmd file.
The second way is to download the data from the GoogleSheet into SKY. In order to do so, you have to use the gsheet package, by calling it into an R code chunk with library(gsheet). I generally put all calls to packages in the first R code chunk of the RMarkdown document, that I generally call libraries, as I did in the GrasslandPES.Rmd example file. Then you simply have to call the data from GoogleSheet using the gsheet2tbl command: data <- gsheet2tbl("YourGoogleSheetUrl"). See the example in the code chunk sendingGCP in the GrasslandPES.Rmd file.
The third way is to download data from your computer. Here is a quick tutorial on how to do that for different file formats. In order to make the data really available to the community though, you have to either put it into a GoogleSheet or send it to the MySQL repo (see below).
The fourth way is to send the data directly to the SKY MySQL repo. In order to do so, you need to download the RMySQL package by adding library(RMySQL) to the libraries code chunk. Then, you need to connect to the SKY MySQL database. For this, you have to include the following code chunk in your page (I suggest to place it right after the library chunk):
```{r source,echo=FALSE,results=FALSE,warning=FALSE,error=FALSE,message=FALSE}
source('source.R')
read_chunk('SKY_core_functions.R')
SKY.db <- dbConnect(MySQL(),dbname=dbname,username=username,password=password,host=host)
`````
If you are sending a full table to the MySQL repo, you can use the following command to send the R dataframe called data to the SKY database, under the name NewTable: dbWriteTable(SKY.db,'NewTable',data,overwrite=TRUE). See the example in the code chunk sendingGCP in the GrasslandPES.Rmd file.
If you want to update an already existing Table in the SKY MySQL database, use the dbUpdateResults function that I’ve written. Imagine you want to send results stored in the variables estimate1 and estimate2 of an R dataframe called data to a table in the SKY MySQL server called Table, and that you want to update in this table only the entries that have the same characteristics where1 and where2 than in your dataframe (for example same program, or same paper id). Just call: dbUpdateResults(con=SKY.db,table='Table',data=data,where=c('where1','where2'),update=c('estimate1','estimate2')). See the example in the code chunk sending.meta in the GrasslandPES.Rmd file.
Run SKY with your changes
In order to run the changes you have made to SKY to see if they work, I suggest a two-step approach, but feel free to skip step one.
- Check that the page you have modified runs as a stand alone document. In order to do so, click on the
knitbutton in the top left corner of the Rstudio window.

A new html page should appear. How nice, right? 2. Run the whole SKY website. For that, hit the Build Website button, yes, the one with a nice hammer on it.

OK, does the nice SKY website appear? If not, then you might have a bug.
Debugging
Debugging in Rstudio is made super easy in this tutorial. You have to run the R code alone in order to debug, which is made super easy by the Run button in Rstudio, on the top right corner of the main window, the one with a green arrow on it.
Bug messages from RMarkdown are in general pretty useful, so you should be able to decide where the problem comes from (R or Markdown) and from which chunk or part of the text.
Uploading your changes to SKY
Ok, so now you’ve worked on SKY for some time, you have made adjustments, corrected typos, tinkered with the code, maybe even added a page or some data. How do you move all of this to SKY? This is where Git and GitHub come in handy Let’s read on ;)
Commiting your changes to Git
The first thing to do is to tell your local Git repository that you have made all these changes. In order to do so, click on the Git thumbnail on the top right corner of the Rstudio window.
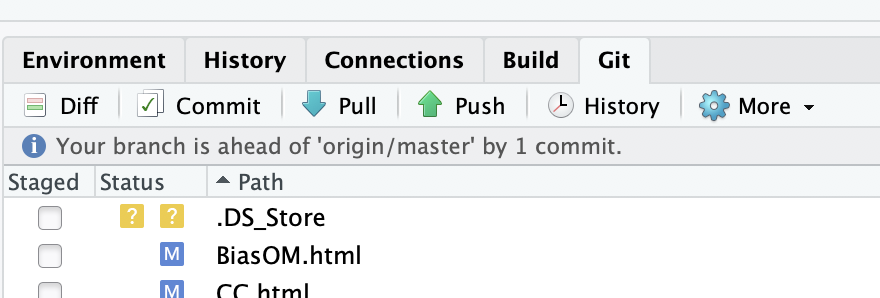
All the files that you have either created or modified should appear there. The files that you have modified should have a blue M in front of them. The files that you have created should have a yellow ? in front of them.
Click on the Staged button in front of each of the modified or newly created file. This stages your files for Git. For a modified file, it tells Git that it should commit their changes if you ask him to. The blue M moves to the left, indicating that the modifications are now ready to be committed. For a created file, staging says to Git to add them to the list of files that it follows. A green A appears in front of the file, it has been Added to the Git repo.
Click on the Commit button at the top of the Git window. A new window, the committing window, appears.

Type a clear commit message explaining what you did in the Commit message box. Click on the Commit button just below the Commit message box. A new box opens. Close it when the Git code has finished running. Close the commit window. Done!
Sending your changes to your GitHub repository
You will not believe this, but sending your changes to GitHub repository only requires to click on one button. Yes, only one. It is the Push button marked with a green arrow pointing upwards in the Git panel.
Go ahead and click ;) A box should open, and then a window asking you your id and password for accessing your GitHub account. Fill in the info. Click on Submit. Done! You have almost made your first contribution to open science.
Sending a merge request
When you are happy with what you have done and do not see how to improve it any further, you can notify SKY of your changes and ask for them to be included into the actual website. In order to do so, you have to log in to your GitHub account and go to the SKY page on YOUR account https://github.com/YourGitHubUserName/SKY. Click on the New Pull Request button and follow the instructions that you are given.
If you are uncertain of what to do, look here. Once you have submitted your pull request, I will examine it, we might have some back and forth discussion and then, when everything is OK, I’ll accept the pull request. Your work will be merged into SKY and will appear online. You will have made your first contribution to open science! Congrats :)